Genetic Variant Classifications
- by user1
- 01 March, 2022
Predict whether a variant will have conflicting clinical classifications.
LicenseCC0: Public Domain
Tagsearth and nature, biology, classification, healthcare, medicineand 1 more
Context
ClinVar is a public resource containing annotations about human genetic variants. These variants are (usually manually) classified by clinical laboratories on a categorical spectrum ranging from benign, likely benign, uncertain significance, likely pathogenic, and pathogenic. Variants that have conflicting classifications (from laboratory to laboratory) can cause confusion when clinicians or researchers try to interpret whether the variant has an impact on the disease of a given patient.
Content
The objective is to predict whether a ClinVar variant will have conflicting classifications. This is presented here as a binary classification problem, where each record in the dataset is a genetic variant.
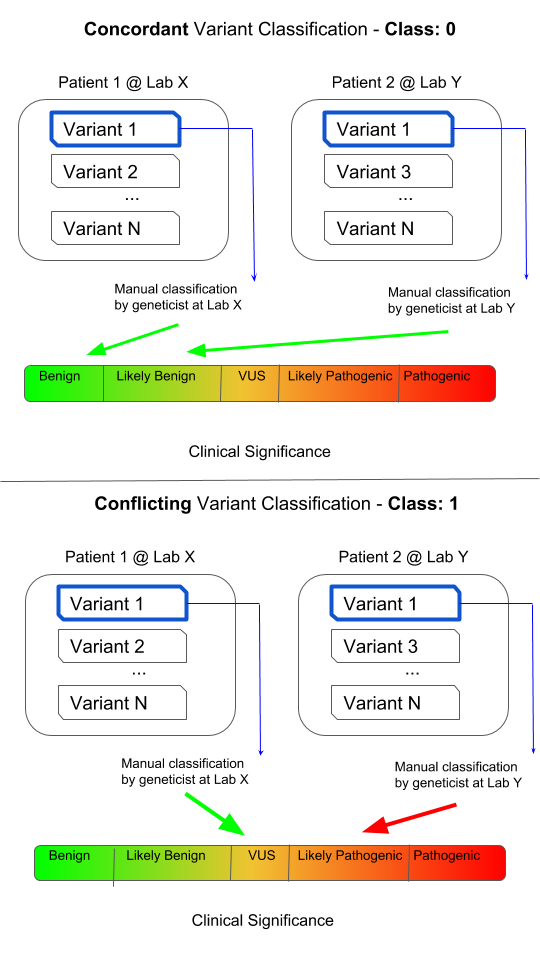
Conflicting classifications are when two of any of the following three categories are present for one variant, two submissions of one category are not considered conflicting.
- Likely Benign or Benign
- VUS
- Likely Pathogenic or Pathogenic
Conflicting classification has been assigned to the CLASS column. It is a binary representation of whether or not a variant has conflicting classifications, where 0 represents consistent classifications and 1 represents conflicting classifications.
Since this problem only relates to variants with multiple classifications, I removed all variants from the original ClinVar .vcf which only had one submission.
The raw variant call format (vcf) file was downloaded here on Saturday, April 7th, 2018:
ftp://ftp.ncbi.nlm.nih.gov/pub/clinvar/vcf_GRCh37/clinvar.vcf.gz
Scripts used to generate this file in this repo
Acknowledgements
Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Jang W, Karapetyan K, Katz K, Liu C, Maddipatla Z, Malheiro A, McDaniel K, Ovetsky M, Riley G, Zhou G, Holmes JB, Kattman BL, Maglott DR. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018 Jan 4. PubMed PMID: 29165669.
Inspiration
I’m exploring ideas for applying machine learning to genomics. I’m hoping this dataset will encourage others to think about the additional feature engineering that’s necessary to confidently assess the objective. There could be a benefit to identifying single submission variants that may yet to have assigned a conflicting classification.
Size: 3673 KB Price: Free Author: Kevin Arvai and 1 collaborator Data source: kaggle.com